Ethylene: A Master Regulator of Plant–Microbe Interactions under Abiotic Stresses
Abstract
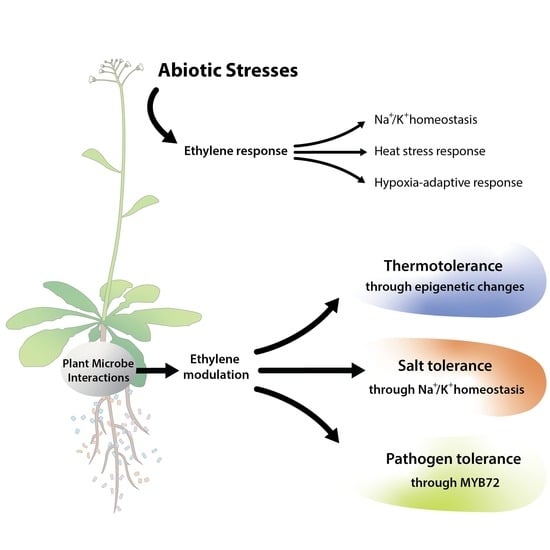
:1. Introduction
2. Ethylene and Plant Abiotic Stresses
2.1. Salt Stress
2.2. Hypoxia
2.3. Heat Stress
3. Ethylene and Plant–Microbe Interactions
4. Ethylene and Pathogenic Plant–Microbe Interactions
5. Ethylene and Beneficial Plant–Microbe Interactions
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rodriguez, P.A.; Rothballer, M.; Chowdhury, S.P.; Nussbaumer, T.; Gutjahr, C.; Falter-Braun, P. Systems Biology of Plant-Microbiome Interactions. Mol. Plant 2019, 12, 804–821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malgioglio, G.; Rizzo, G.F.; Nigro, S.; du Prey, V.L.; Herforth-Rahmé, J.; Catara, V.; Branca, F. Plant-Microbe Interaction in Sustainable Agriculture: The Factors That May Influence the Efficacy of PGPM Application. Sustainability 2022, 14, 2253. [Google Scholar] [CrossRef]
- Pierik, R.; Tholen, D.; Poorter, H.; Visser, E.J.; Voesenek, L.A. The Janus face of ethylene: Growth inhibition and stimulation. Trends Plant Sci. 2006, 11, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Ravanbakhsh, M.; Sasidharan, R.; Voesenek, L.A.C.J.; Kowalchuk, G.A.; Jousset, A. Microbial modulation of plant ethylene signaling: Ecological and evolutionary consequences. Microbiome 2018, 6, 52. [Google Scholar] [CrossRef] [PubMed]
- Schaller, G.E. Ethylene and the regulation of plant development. BMC Biol. 2012, 10, 9. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Khan, M.Y.; Carvalhais, L.C.; Delgado-Baquerizo, M.; Yan, L.; Crawford, M.; Dennis, P.G.; Singh, B.; Schenk, P.M. Soil amendments with ethylene precursor alleviate negative impacts of salinity on soil microbial properties and productivity. Sci. Rep. 2019, 9, 6892. [Google Scholar] [CrossRef] [Green Version]
- Arraes, F.B.M.; Beneventi, M.A.; De Sa, M.E.L.; Paixao, J.F.R.; Albuquerque, E.V.S.; Marin, S.R.R.; Purgatto, E.; Nepomuceno, A.L.; Grossi-De-Sa, M.F. Implications of ethylene biosynthesis and signaling in soybean drought stress tolerance. BMC Plant Biol. 2015, 15, 213. [Google Scholar] [CrossRef]
- Sun, X.; Zhao, T.; Gan, S.; Ren, X.; Fang, L.; Karungo, S.K.; Wang, Y.; Chen, L.; Li, S.; Xin, H. Ethylene positively regulates cold tolerance in grapevine by modulating the expression of ETHYLENE RESPONSE FACTOR 057. Sci. Rep. 2016, 6, 24066. [Google Scholar] [CrossRef] [Green Version]
- Abeles, F.B.; Morgan, P.W.; Saltveit, M.E., Jr. Ethylene in Plant Biology, 2nd ed.; Academic Press: San Diego, CA, USA, 1992. [Google Scholar]
- He, M.-W.; Wang, Y.; Wu, J.-Q.; Shu, S.; Sun, J.; Guo, S.-R. Isolation and characterization of S-Adenosylmethionine synthase gene from cucumber and responsive to abiotic stress. Plant Physiol. Biochem. 2019, 141, 431–445. [Google Scholar] [CrossRef]
- Pattyn, J.; Vaughan-Hirsch, J.; Van De Poel, B. The regulation of ethylene biosynthesis: A complex multilevel control circuitry. New Phytol. 2021, 229, 770–782. [Google Scholar] [CrossRef]
- Chen, Y.-F.; Gao, Z.; Kerris, R.J.; Wang, W.; Binder, B.M.; Schaller, G.E. Ethylene Receptors Function as Components of High-Molecular-Mass Protein Complexes in Arabidopsis. PLoS ONE 2010, 5, e8640. [Google Scholar] [CrossRef] [PubMed]
- Shakeel, S.N.; Gao, Z.; Amir, M.; Chen, Y.-F.; Rai, M.I.; Haq, N.U.; Schaller, G.E. Ethylene Regulates Levels of Ethylene Receptor/CTR1 Signaling Complexes in Arabidopsis thaliana. J. Biol. Chem. 2015, 290, 12415–12424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, H.; Yin, C.; Ma, B.; Chen, S.; Zhang, J. Ethylene signaling in rice and Arabidopsis: New regulators and mechanisms. J. Integr. Plant Biol. 2021, 63, 102–125. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, F.X.; Rossi, M.J.; Glick, B.R.; Nascimento, F.X.; Rossi, M.J.; Glick, B.R. Ethylene and 1-Aminocyclopropane-1-carboxylate (ACC) in Plant–Bacterial Interactions. Front. Plant Sci. 2018, 9, 114. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Ullah, F.; Zhou, D.-X.; Yi, M.; Zhao, Y. Mechanisms of ROS Regulation of Plant Development and Stress Responses. Front. Plant Sci. 2019, 10, 800. [Google Scholar] [CrossRef] [PubMed]
- Ruzicka, K.; Ljung, K.; Vanneste, S.; Podhorská, R.; Beeckman, T.; Friml, J.; Benková, E. Ethylene Regulates Root Growth through Effects on Auxin Biosynthesis and Transport-Dependent Auxin Distribution. Plant Cell 2007, 19, 2197–2212. [Google Scholar] [CrossRef] [Green Version]
- Vaseva, I.I.; Qudeimat, E.; Potuschak, T.; Du, Y.; Genschik, P.; Vandenbussche, F.; Van Der Straeten, D. The plant hormone ethylene restricts Arabidopsis growth via the epidermis. Proc. Natl. Acad. Sci. USA 2018, 115, E4130–E4139. [Google Scholar] [CrossRef] [Green Version]
- Street, I.H.; Aman, S.; Zubo, Y.; Ramzan, A.; Wang, X.; Shakeel, S.N.; Kieber, J.J.; Schaller, G.E. Ethylene Inhibits Cell Proliferation of the Arabidopsis Root Meristem. Plant Physiol. 2015, 169, 338–350. [Google Scholar] [CrossRef] [Green Version]
- Berkowitz, O.; Xu, Y.; Liew, L.C.; Wang, Y.; Zhu, Y.; Hurgobin, B.; Lewsey, M.G.; Whelan, J. RNA-seq analysis of laser microdissected Arabidopsis thaliana leaf epidermis, mesophyll and vasculature defines tissue-specific transcriptional responses to multiple stress treatments. Plant J. 2021, 107, 938–955. [Google Scholar] [CrossRef]
- Eichmann, R.; Richards, L.; Schäfer, P. Hormones as go-betweens in plant microbiome assembly. Plant J. 2021, 105, 518–541. [Google Scholar] [CrossRef]
- Glick, B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014, 169, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Pandey, S. ACC Deaminase Producing Bacteria with Multifarious Plant Growth Promoting Traits Alleviates Salinity Stress in French Bean (Phaseolus vulgaris) Plants. Front. Microbiol. 2019, 10, 1506. [Google Scholar] [CrossRef] [PubMed]
- Camehl, I.; Sherameti, I.; Venus, Y.; Bethke, G.; Varma, A.; Lee, J.; Oelmüller, R. Ethylene signalling and ethylene-targeted transcription factors are required to balance beneficial and nonbeneficial traits in the symbiosis between the endophytic fungus Piriformospora indica and Arabidopsis thaliana. New Phytol. 2010, 185, 1062–1073. [Google Scholar] [CrossRef] [PubMed]
- Khatabi, B.; Schäfer, P. Ethylene in mutualistic symbioses. Plant Signal. Behav. 2012, 7, 1634–1638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, F.; Zhang, F. Cell Cycle Regulation in the Plant Response to Stress. Front. Plant Sci. 2020, 10, 1765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- dos Santos, T.B.; Ribas, A.F.; de Souza, S.G.H.; Budzinski, I.G.F.; Domingues, D.S. Physiological Responses to Drought, Salinity, and Heat Stress in Plants: A Review. Stresses 2022, 2, 113–135. [Google Scholar] [CrossRef]
- Ullah, A.; Bano, A.; Khan, N. Climate Change and Salinity Effects on Crops and Chemical Communication Between Plants and Plant Growth-Promoting Microorganisms Under Stress. Front. Sustain. Food Syst. 2021, 5, 161. [Google Scholar] [CrossRef]
- EL Sabagh, A.; Islam, M.S.; Skalicky, M.; Raza, M.A.; Singh, K.; Hossain, M.A.; Hossain, A.; Mahboob, W.; Iqbal, M.A.; Ratnasekera, D.; et al. Salinity Stress in Wheat (Triticum aestivum L.) in the Changing Climate: Adaptation and Management Strategies. Front. Agron. 2021, 3, 648694. [Google Scholar] [CrossRef]
- Dou, L.; He, K.; Higaki, T.; Wang, X.; Mao, T. Ethylene signaling modulates cortical microtubule reassembly in response to salinity stress. Plant Physiol. 2018, 176, 2071–2081. [Google Scholar] [CrossRef]
- Cheng, Z.; Zhang, X.; Zhao, K.; Yao, W.; Li, R.; Zhou, B.; Jiang, T. Over-Expression of ERF38 Gene Enhances Salt and Osmotic Tolerance in Transgenic Poplar. Front. Plant Sci. 2019, 10, 1375. [Google Scholar] [CrossRef]
- Riyazuddin, R.; Verma, R.; Singh, K.; Nisha, N.; Keisham, M.; Bhati, K.; Kim, S.; Gupta, R. Ethylene: A Master Regulator of Salinity Stress Tolerance in Plants. Biomolecules 2020, 10, 959. [Google Scholar] [CrossRef] [PubMed]
- Freitas, V.S.; Miranda, R.D.S.; Costa, J.H.; de Oliveira, D.F.; Paula, S.D.O.; Miguel, E.D.C.; Freire, R.S.; Prisco, J.T.; Gomes-Filho, E. Ethylene triggers salt tolerance in maize genotypes by modulating polyamine catabolism enzymes associated with H2O2 production. Environ. Exp. Bot. 2018, 145, 75–86. [Google Scholar] [CrossRef]
- Xu, L.; Xiang, G.; Sun, Q.; Ni, Y.; Jin, Z.; Gao, S.; Yao, Y. Melatonin enhances salt tolerance by promoting MYB108A-mediated ethylene biosynthesis in grapevines. Hortic. Res. 2019, 6, 114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takács, Z.; Czékus, Z.; Tari, I.; Poór, P. The role of ethylene signalling in the regulation of salt stress response in mature tomato fruits: Metabolism of antioxidants and polyamines. J. Plant Physiol. 2022, 277, 153793. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zu, Y.-G.; Tang, Z.-H. Ethylene improves Arabidopsis salt tolerance mainly via retaining K+ in shoots and roots rather than decreasing tissue Na+ content. Environ. Exp. Bot. 2013, 86, 60–69. [Google Scholar] [CrossRef]
- Assaha, D.V.M.; Ueda, A.; Saneoka, H.; Al-Yahyai, R.; Yaish, M.W. The role of Na+ and K+ transporters in salt stress adaptation in glycophytes. Front. Physiol. 2017, 8, 509. [Google Scholar] [CrossRef] [Green Version]
- Lang, T.; Deng, C.; Yao, J.; Zhang, H.; Wang, Y.; Deng, S. A Salt-Signaling Network Involving Ethylene, Extracellular ATP, Hydrogen Peroxide, and Calcium Mediates K+/Na+ Homeostasis in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 8683. [Google Scholar] [CrossRef] [PubMed]
- Sehar, Z.; Iqbal, N.; Khan, M.I.R.; Masood, A.; Rehman, T.; Hussain, A.; AlAjmi, M.F.; Ahmad, A.; Khan, N.A. Ethylene reduces glucose sensitivity and reverses photosynthetic repression through optimization of glutathione production in salt-stressed wheat (Triticum aestivum L.). Sci. Rep. 2021, 11, 12650. [Google Scholar] [CrossRef]
- Iqbal, N.; Nazar, R.; Khan, M.I.R.; Khan, N.A. Variation in photosynthesis and growth of mustard cultivars: Role of ethylene sensitivity. Sci. Hortic. 2012, 135, 1–6. [Google Scholar] [CrossRef]
- Khan, N.A. An evaluation of the effects of exogenous ethephon, an ethylene releasing compound, on photosynthesis of mustard (Brassica juncea) cultivars that difer in photosynthetic capacity. BMC Plant Biol. 2004, 4, 21. [Google Scholar] [CrossRef]
- Iqbal, N.; Nazar, R.; Syeed, S.; Masood, A.; Khan, N.A. Exogenously-sourced ethylene increases stomatal conductance, photosynthesis, and growth under optimal and deficient nitrogen fertilization in mustard. J. Exp. Bot. 2011, 62, 4955–4963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loreti, E.; Perata, P. The Many Facets of Hypoxia in Plants. Plants 2020, 9, 745. [Google Scholar] [CrossRef] [PubMed]
- León, J.; Castillo, M.C.; Gayubas, B. The hypoxia–reoxygenation stress in plants. J. Exp. Bot. 2020, 72, 5841–5856. [Google Scholar] [CrossRef]
- Hartman, S.; Sasidharan, R.; Voesenek, L.A.C.J. The role of ethylene in metabolic acclimations to low oxygen. New Phytol. 2019, 229, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, D.J.; Lee, S.C.; Isa, N.M.; Gramuglia, S.; Fukao, T.; Bassel, G.W.; Correia, C.S.; Corbineau, F.; Theodoulou, F.L.; Bai-ley-Serres, J.; et al. Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 2021, 479, 415–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Licausi, F.; van Dongen, J.T.; Giuntoli, B.; Novi, G.; Santaniello, A.; Geigenberger, P.; Perata, P. HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J. 2010, 62, 302–315. [Google Scholar] [CrossRef]
- Weits, D.A.; Giuntoli, B.; Kosmacz, M.; Parlanti, S.; Hubberten, H.M.; Riegler, H.; Hoefgen, R.; Perata, P.; van Dongen, J.T.; Licausi, F. Plant cysteine oxidases control the oxygen-dependent branch of the N-end-rule pathway. Nat. Commun. 2014, 5, 3425. [Google Scholar] [CrossRef] [Green Version]
- Gibbs, D.J.; Md Isa, N.; Movahedi, M.; Lozano-Juste, J.; Mendiondo, G.M.; Berckhan, S.; Marín-de la Rosa, N.; Vicente Conde, J.; Sousa Correia, C.; Pearce, S.P.; et al. Nitric oxide sensing in plants is mediated by pro-teolytic control of group VII ERF transcription factors. Mol. Cell. 2014, 53, 369–379. [Google Scholar] [CrossRef] [Green Version]
- Gibbs, D.J.; Tedds, H.M.; Labandera, A.M.; Bailey, M.; White, M.D.; Hartman, S.; Sprigg, C.; Mogg, S.L.; Osborne, R.; Dambire, C.; et al. Oxygen-dependent proteolysis regulates the stability of angiosperm polycomb repressive complex 2 subunit VERNALIZATION 2. Nat. Commun. 2018, 9, 5438. [Google Scholar] [CrossRef] [Green Version]
- Weits, D.A.; Kunkowska, A.B.; Kamps, N.C.W.; Portz, K.M.S.; Packbier, N.K.; Nemec Venza, Z.; Gaillochet, C.; Lohmann, J.U.; Pedersen, O.; van Dongen, J.T.; et al. An apical hypoxic niche sets the pace of shoot meristem activity. Nature 2019, 569, 714–717. [Google Scholar] [CrossRef]
- Morrell, S.; Greenway, H. Evidence does not support ethylene as a cue for synthesis of alcohol dehydrogenase and pyruvate decarboxylase during exposure to hypoxia. Funct. Plant Biol. 1989, 16, 469–475. [Google Scholar] [CrossRef]
- Peng, H.P. Signaling events in the hypoxic induction of alcohol dehydrogenase gene in Arabidopsis. Plant Physiol. 2001, 126, 742–749. [Google Scholar] [CrossRef] [Green Version]
- van Veen, H.; Mustroph, A.; Barding, G.A.; Eijk, M.V.; Welschen-Evertman, R.A.M.; Pedersen, O.; Visser, E.J.W.; Larive, C.K.; Pierik, R.; Bailey-Serres, J.; et al. Two Rumex species from contrasting hydrological niches regulate flooding tolerance through distinct mechanisms. Plant Cell. 2013, 25, 4691–4707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotak, S.; Larkindale, J.; Lee, U.; von Koskull-Döring, P.; Vierling, E.; Scharf, K.D. Complexity of the heat stress response in plants. Curr. Opin. Plant Biol. 2007, 10, 310–316. [Google Scholar]
- Lamke, J.; Brzezinka, K.; Altmann, S.; Bäurle, I. A hit-and-run heat shock factor governs sustained histone methylation and transcriptional stress memory. EMBO J. 2016, 35, 162–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shekhawat, K.; Almeida-Trapp, M.; García-Ramírez, G.X.; Hirt, H. Beat the heat: Plant- and microbe-mediated strategies for crop thermotolerance. Trends Plant Sci. 2022, 27, 802–813. [Google Scholar] [CrossRef] [PubMed]
- Jegadeesan, S.; Chaturvedi, P.; Ghatak, A.; Pressman, E.; Meir, S.; Faigenboim, A.; Rutley, N.; Beery, A.; Harel, A.; Weckwerth, W.; et al. Proteomics of Heat-Stress and Ethylene-Mediated Thermotolerance Mechanisms in Tomato Pollen Grains. Front. Plant Sci. 2018, 9, 1558. [Google Scholar] [CrossRef] [Green Version]
- Sehar, Z.; Gautam, H.; Iqbal, N.; Alvi, A.F.; Jahan, B.; Fatma, M.; Albaqami, M.; Khan, N.A. The Functional Interplay between Ethylene, Hydrogen Sulfide, and Sulfur in Plant Heat Stress Tolerance. Biomolecules 2022, 12, 678. [Google Scholar] [CrossRef]
- Wu, Y.-S.; Yang, C.-Y. Ethylene-mediated signaling confers thermotolerance and regulates transcript levels of heat shock factors in rice seedlings under heat stress. Bot. Stud. 2019, 60, 23. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Zhao, X.; Bürger, M.; Wang, Y.; Chory, J. Two interacting ethylene response factors regulate heat stress response. Plant Cell. 2021, 33, 338–357. [Google Scholar] [CrossRef]
- Shekhawat, K.; Saad, M.M.; Sheikh, A.; Mariappan, K.; Al-Mahmoudi, H.; Abdulhakim, F.; Eida, A.A.; Masmoudi, K.J.; Hirt, H. Root endophyte induced plant thermotolerance by constitutive chromatin modification at heat stress memory gene loci. EMBO Rep. 2021, 22, e51049. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, P.J.P.; Colaianni, N.R.; Fitzpatrick, C.R.; Dangl, J.L. Beyond pathogens: Microbiota interactions with the plant immune system. Curr. Opin. Microbiol. 2019, 49, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Broekaert, W.F.; Delauré, S.L.; De Bolle, M.F.; Cammue, B.P. The Role of Ethylene in Host-Pathogen Interactions. Annu. Rev. Phytopathol. 2006, 44, 393–416. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, N.; Khan, N.A.; Ferrante, A.; Trivellini, A.; Francini, A.; Khan, M.I.R. Ethylene Role in Plant Growth, Development and Senescence: Interaction with Other Phytohormones. Front. Plant Sci. 2017, 08, 475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pré, M.; Atallah, M.; Champion, A.; De Vos, M.; Pieterse, C.M.J.; Memelink, J. The AP2/ERF Domain Transcription Factor ORA59 Integrates Jasmonic Acid and Ethylene Signals in Plant Defense. Plant Physiol. 2008, 147, 1347–1357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solano, R.; Stepanova, A.; Chao, Q.; Ecker, J.R. Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 1998, 12, 3703–3714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boutrot, F.; Segonzac, C.; Chang, K.N.; Qiao, H.; Ecker, J.R.; Zipfel, C.; Rathjen, J.P. Direct transcriptional control of the Arabidopsis immune receptor FLS2 by the ethylene-dependent transcription factors EIN3 and EIL1. Proc. Natl. Acad. Sci. USA 2010, 107, 14502–14507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berrocal-Lobo, M.; Molina, A.; Solano, R. Constitutive expression of ETHYLENERESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 2002, 29, 23–32. [Google Scholar] [CrossRef]
- Spoel, S.H.; Johnson, J.S.; Dong, X. Regulation of tradeoffs between plant defenses against pathogens with different life-styles. Proc Natl Acad Sci USA. 2007, 104, 18842–18847. [Google Scholar] [CrossRef] [Green Version]
- Verhage, A.; Vlaardingerbroek, I.; Raaijmakers, C.; Van Dam, N.; Dicke, M.; Van Wees, S.C.M.; Pieterse, C.M. Rewiring of the jasmonate signaling pathway in Arabidopsis during insect herbivory. Front. Plant Sci. 2011, 2, 47. [Google Scholar] [CrossRef] [Green Version]
- Vleesschauwer, D.E.; Exu, J.; Hã¶fte, M. Making sense of hormone-mediated defense networking: From rice to Arabidopsis. Front. Plant Sci. 2014, 5, 611. [Google Scholar] [CrossRef] [PubMed]
- Huckelhoven, R.; Fodor, J.; Preis, C.; Kogel, K.H. Hypersensitive cell death and papilla formation in barley attacked by the powdery mildew fungus are associated with hydrogen peroxide but not with salicylic acid accumulation. Plant Physiol. 1999, 119, 1251–1260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glazebrook, J. Contrasting Mechanisms of Defense Against Biotrophic and Necrotrophic Pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef] [PubMed]
- Leon-Reyes, A.; Spoel, S.H.; De Lange, E.S.; Abe, H.; Kobayashi, M.; Tsuda, S.; Millenaar, F.F.; Welschen, R.A.M.; Ritsema, T.; Pieterse, C.M.J. Ethylene modulates the role of NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 in cross talk between salicylate and jasmonate signaling. Plant Physiol. 2009, 149, 1797–1809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leon-Reyes, A.; Du, Y.; Koornneef, A.; Proietti, S.; Körbes, A.P.; Memelink, J.; Pieterse, C.M.J.; Ritsema, T. Ethylene signaling renders the jasmonate response of Arabidopsis insensitive to future suppression by salicylic Acid. Mol. Plant Mi-crobe Interact. 2010, 23, 187–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zander, M.; La Camera, S.; Lamotte, O.; Métraux, J.P.; Gatz, C. Arabidopsis thaliana class-II TGA transcription factors are essential activators of jasmonic acid/ethylene-induced defense responses. Plant J. 2010, 61, 200–210. [Google Scholar] [CrossRef] [Green Version]
- Lorenzo, O.; Piqueras, R.; Sánchez-Serrano, J.J.; Solano, R. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell. 2003, 15, 165–178. [Google Scholar] [CrossRef] [Green Version]
- Kolomiets, M.V.; Chen, H.; Gladon, R.J.; Braun, E.; Hannapel, D.J. A Leaf Lipoxygenase of Potato Induced Specifically by Pathogen Infection. Plant Physiol. 2000, 124, 1121–1130. [Google Scholar] [CrossRef] [Green Version]
- Kondo, S.; Yamada, H.; Setha, S. Effect of jasmonates differed at fruit ripening stages on 1-aminocyclopropane-1-carboxylate (ACC) synthase and ACC oxidase gene expression in pears. J. Am. Soc. Hort. Sci. 2007, 132, 120–125. [Google Scholar] [CrossRef] [Green Version]
- Epple, P.; Bohlmann, K.P.H. An Arabidopsis thaliana thionin gene is inducible via a signal transduction pathway different from that for pathogenesis-related proteins. Plant Physiol. 1995, 109, 813–820. [Google Scholar] [CrossRef] [Green Version]
- Norman-Setterbald, C.; Vidal, S.; Palva, E.T. Interaction signal pathways control defense gene expression in Arabidopsis in response to cell wall-degrading enzymes from Erwinia carotovora. Mol. Plant Microbe Interact. 2000, 13, 430–438. [Google Scholar] [CrossRef] [PubMed]
- Doares, S.H.; Narváez-Vásquex, J.; Conconi, A.; Ryan, C.A. Salicylic acid inhibits synthesis of proteinase inhibitors in to-mato leaves induced by systemin and jasmonic acid. Plant Physiol. 1995, 108, 1741–1746. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, P.; Buchala, A.; Métraux, J.P. Gene-expression patterns and levels of jasmonic acid in rice treated with the re-sistance inducer 2,6-dichloroisonicotinic acid. Plant Physiol. 1997, 115, 61–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, P.; Dong, Z.; Guo, P.; Zhang, X.; Qiu, Y.; Li, B.; Wang, Y.; Guo, H. Salicylic Acid Suppresses Apical Hook Formation via NPR1-Mediated Repression of EIN3 and EIL1 in Arabidopsis. Plant Cell 2019, 32, 612–629. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Xue, L.; Chintamanani, S.; Germain, H.; Lin, H.; Cui, H.; Cai, R.; Zuo, J.; Tang, X.; Li, X.; et al. ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFI-CIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell. 2009, 21, 2527–2540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, A.; Singh, S.; Gaurav, A.K.; Srivastava, S.; Verma, J.P. Plant Growth-Promoting Bacteria: Biological Tools for the Mitigation of Salinity Stress in Plants. Front. Microbiol. 2020, 11, 1216. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R. Stress control and ACC deaminase. In Principles of Plant-Microbe Interactions; Springer: Cham, Switzerland, 2015; pp. 257–264. [Google Scholar]
- Kumar, M.; Giri, V.P.; Pandey, S.; Gupta, A.; Patel, M.K.; Bajpai, A.B.; Jenkins, S.; Siddique, K.H.M. Plant-Growth-Promoting Rhizobacteria emerging as an effective bioinoculant to improve the growth, production, and stress tolerance of vegetable crops. Int. J. Mol. Sci. 2021, 22, 12245. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Mishra, R.; Rai, S.; Bano, A.; Pathak, N.; Fujita, M.; Kumar, M.; Hasanuzzaman, M. Mechanistic Insights of Plant Growth Promoting Bacteria Mediated Drought and Salt Stress Tolerance in Plants for Sustainable Agriculture. Int. J. Mol. Sci. 2022, 23, 3741. [Google Scholar] [CrossRef]
- Soni, R.; Yadav, S.K.; Rajput, A.S. ACC-Deaminase Producing Rhizobacteria: Prospects and Application as Stress Busters for Stressed Agriculture. In Microorganisms for Sustainability Book Series; MICRO: Singapore, 2018; Volume 7. [Google Scholar] [CrossRef]
- Chandra, D.; Srivastava, R.; Gupta, V.V.S.R.; Franco, C.M.M.; Sharma, A.K. Evaluation of ACC-deaminase-producing rhizobacteria to alleviate water-stress impacts in wheat (Triticum aestivum L.) plants. Can. J. Microbiol. 2019, 65, 387–403. [Google Scholar] [CrossRef]
- Danish, S.; Zafar-Ul-Hye, M.; Mohsin, F.; Hussain, M. ACC-deaminase producing plant growth promoting rhizobacteria and biochar mitigate adverse effects of drought stress on maize growth. PLoS ONE 2020, 15, e0230615. [Google Scholar] [CrossRef]
- Nadeem, S.M.; Zahir, Z.A.; Naveed, M. Rhizobacteria containing ACC-deaminase confer salt tolerance in maize grown on salt-affected fields. Can. J. Microbiol. 2009, 55, 1302–1309. [Google Scholar] [CrossRef] [PubMed]
- Akram, W.; Aslam, H.; Ahmad, S.R.; Anjum, T.; Yasin, N.A.; Khan, W.U.; Ahmad, A.; Guo, J.; Wu, T.; Luo, W.; et al. Bacillus megaterium strain A12 ameliorates salinity stress in tomato plants through multiple mechanisms. J. Plant Interact. 2019, 14, 506–518. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, H.M.; Fiaz, S.; Hafeez, S.; Zahra, S.; Shah, A.N.; Gul, B.; Aziz, O.; Rahman, M.U.; Fakhar, A.; Rafique, M.; et al. Plant Growth-Promoting Rhizobacteria Eliminate the Effect of Drought Stress in Plants: A Review. Front. Plant Sci. 2022, 13, 1965. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Charles, T.; Glick, B.R. Amelioration of high salinity stress damage by plant growth-promoting bacterial endophytes that contain ACC deaminase. Plant Physiol. Biochem. 2014, 80, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wang, R.; Yang, Z.; Zhan, Y.; Ma, Y.; Ping, S.; Zhang, L.; Lin, M.; Yan, Y. 1-Aminocyclopropane-1-Carboxylate Deaminase from Pseudomonas stutzeri A1501 Facilitates the Growth of Rice in the Presence of Salt or Heavy Metals. J. Microbiol. Biotechnol. 2015, 25, 1119–1128. [Google Scholar] [CrossRef]
- Shaharoona, B.; Arshad, M.; Zahir, Z.A. Effect of plant growth promoting rhizobacteria containing ACC-deaminase on maize (Zea mays L.) growth under axenic conditions and on nodulation in mung bean (Vigna radiata L.). Lett. Appl Mi-crobiol. 2006, 42, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Peskan-Berghofer, T.; Shahollari, B.; Giong, P.H.; Hehl, S.; Markert, C.; Blanke, V.; Kost, G.; Varma, A.; Oelmuller, R. Association of Piriformospora indica with Arabidopsis thaliana roots represents a novel system to study beneficial plant-microbe interactions and involves early plant protein modifications in the endoplasmic reticulum and at the plasma membrane. Physiol. Plant. 2004, 122, 465–477. [Google Scholar] [CrossRef]
- Oldroyd, G.E.D.; Engstrom, E.M.; Long, S.R. Ethylene Inhibits the Nod Factor Signal Transduction Pathway of Medicago truncatula. Plant Cell 2001, 13, 1835–1849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geil, R.D.; Peterson, L.R.; Guinel, F.C. Morphological alterations of pea (Pisum sativum cv. Sparkle) arbuscular mycorrhizas as a result of exogenous ethylene treatment. Mycorrhiza 2001, 11, 137–143. [Google Scholar] [CrossRef]
- Enonaka, S.; Eezura, H. Plant–Agrobacterium interaction mediated by ethylene and super-Agrobacterium conferring efficient gene transfer. Front. Plant Sci. 2014, 5, 681. [Google Scholar] [CrossRef]
- Kato, J.; Kim, H.-E.; Takiguchi, N.; Kuroda, A.; Ohtake, H. Pseudomonas aeruginosa as a model microorganism for investigation of chemotactic behaviors in ecosystem. J. Biosci. Bioeng. 2008, 106, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Martín-Rodríguez, J.; León-Morcillo, R.; Vierheilig, H.; Ocampo, J.A.; Ludwig-Müller, J.; García-Garrido, J.M. Ethylene-dependent/ethylene-independent ABA regulation of tomato plants colonized by arbuscular mycorrhiza fungi. New Phytol. 2011, 190, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Plett, J.M.; Khachane, A.; Ouassou, M.; Sundberg, B.; Kohler, A.; Martin, F. Ethylene and jasmonic acid act as negative modulators during mutualistic symbiosis between Laccaria bicolor and Populus roots. New Phytol. 2014, 202, 270–286. [Google Scholar] [CrossRef] [PubMed]
- de Zélicourt, A.; Synek, L.; Saad, M.M.; Alzubaidy, H.; Jalal, R.; Xie, Y.; Andrés-Barrao, C.; Rolli, E.; Guerard, F.; Mariappan, K.G.; et al. Ethylene induced plant stress tolerance by Enterobacter sp. SA187 is mediated by 2-keto-4-methylthiobutyric acid production. PLoS Genet. 2018, 14, e1007273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrés-Barrao, C.; Alzubaidy, H.; Jalal, R.; Mariappan, K.G.; de Zélicourt, A.; Bokhari, A.; Artyukh, O.; Alwutayd, K.; Rawat, A.; Shekhawat, K.; et al. Coordinated bacterial and plant sulfur metabolism in Enterobacter sp. SA187–induced plant salt stress tolerance. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef] [PubMed]
- Datta, R.; Kumar, D.; Sultana, A.; Hazra, S.; Bhattacharyya, D.; Chattopadhyay, S. Glutathione regulates ACC synthase transcription via WRKY33 and ACC oxidase by modulating mRNA stability to induce ethylene synthesis during stress. Plant Physiol. 2015, 169, 2963–2981. [Google Scholar] [CrossRef]
- Poupin, M.J.; Timmermann, T.; Vega, A.; Zuñiga, A.; González, B. Effects of the Plant Growth-Promoting Bacterium Burkholderia phytofirmans PsJN throughout the Life Cycle of Arabidopsis thaliana. PLoS ONE 2013, 8, e69435. [Google Scholar] [CrossRef] [Green Version]
- Zúñiga, A.; Poupin, M.J.; Donoso, R.; Ledger, T.; Guiliani, N.; Gutiérrez, R.A.; González, B. Quorum Sensing and Indole-3-Acetic Acid Degradation Play a Role in Colonization and Plant Growth Promotion of Arabidopsis thaliana by Burkholderia phytofirmans PsJN. Mol. Plant Microbe Interact. 2013, 26, 546–553. [Google Scholar] [CrossRef] [Green Version]
- Pinedo, I.; Ledger, T.; Greve, M.; Poupin, M.J. Burkholderia phytofirmans PsJN induces long-term metabolic and transcriptional changes involved in Arabidopsis thaliana salt tolerance. Front. Plant Sci. 2015, 6, 466. [Google Scholar] [CrossRef] [Green Version]
- Poupin, M.J.; Greve, M.; Carmona, V.; Pinedo, I. A Complex Molecular Interplay of Auxin and Ethylene Signaling Pathways Is Involved in Arabidopsis Growth Promotion by Burkholderia phytofirmans PsJN. Front. Plant Sci. 2016, 7, 492. [Google Scholar] [CrossRef]
- Ton, J.; Davison, S.; Van Wees, S.C.; Van Loon, L.; Pieterse, C.M. The Arabidopsis ISR1 Locus Controlling Rhizobacteria-Mediated Induced Systemic Resistance Is Involved in Ethylene Signaling. Plant Physiol. 2001, 125, 652–661. [Google Scholar] [CrossRef] [Green Version]
- Pieterse, C.M.J.; Van Pelt, J.A.; Ton, J.; Parchmann, S.; Mueller, M.J.; Buchala, A.J.; Métraux, J.P.; Van Loon, L.C. Rhizo-bacteria-mediated induced systemic resistance (ISR) in Arabidopsis requires sensitivity to jasmonate and ethylene but is not accompanied by an increase in their production. Physiol. Mol. Plant Pathol. 2000, 57, 123–134. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shekhawat, K.; Fröhlich, K.; García-Ramírez, G.X.; Trapp, M.A.; Hirt, H. Ethylene: A Master Regulator of Plant–Microbe Interactions under Abiotic Stresses. Cells 2023, 12, 31. https://doi.org/10.3390/cells12010031
Shekhawat K, Fröhlich K, García-Ramírez GX, Trapp MA, Hirt H. Ethylene: A Master Regulator of Plant–Microbe Interactions under Abiotic Stresses. Cells. 2023; 12(1):31. https://doi.org/10.3390/cells12010031
Chicago/Turabian StyleShekhawat, Kirti, Katja Fröhlich, Gabriel X. García-Ramírez, Marilia A. Trapp, and Heribert Hirt. 2023. "Ethylene: A Master Regulator of Plant–Microbe Interactions under Abiotic Stresses" Cells 12, no. 1: 31. https://doi.org/10.3390/cells12010031